
- Bhavin S Khatri
Understanding how evolution works using tools of statistical physics
- Research
 Evolution of Biophysical Genotype Phenotype maps
Evolution of Biophysical Genotype Phenotype mapsUsing analogies from statistical physics to understand emergent phenomena in evolution
Natural selection acts on phenotypes, yet variation in phenotypes arises from mutations at the genetic level. It is being increasingly recognised that a crucial missing ingredient for a full understanding of evolution, even at a qualitative level, is the role of genotype-phenotype maps, or the mapping from sequence to function. Quantitative and predictive theories of evolution that account for this mapping will be required to understand several pressing problems, including antibiotic resistance, virus evolution, and cancer evolution. A key generic prediction is that small drift-dominated populations adapt to phenotypes that have the most sequences coding for them instead of the most optimal (Khatri, PNAS 2009, Genetics 2016). This means smaller populations undergo more rapid speciation as common ancestors are more likely to be maladapted (Khatri, JTB 2015, Khatri, Genetics, 2015 & Khatri, PLoS Computational Biology 2019). In addition, I have developed a stochastic theory for the evolution of phenotypes within a continuous stochastic dynamics framework (Khatri et al, JTB, 2015); this holds promise of greatly simplified modelling of complex genotype-phenotype maps in the future.
 Virus Evolution
Virus EvolutionDetecting selection in longitudinal deep sequencing of viruses
My work aims to understand the basic evolutionary forces that shape the genomes of pathogens and the stochastic dynamics of variants in a host. A key quantity to calculate is the probability of observing a change in the frequency of a variant in a fixed time interval. This is a difficult mathematical diffusion problem that has eluded accurate solution for changes over short times. The key difficulty is the that the variance of allele frequencies (or diffusion constant) is not the same for all allele frequencies. Fisher's angular transformation gives a variance which is independent of a new angular frequency and so effectively simple Brownian motion. The cost is a non-linear potential which is a manifestation of the effects of genetic drift, and is probably why this transformation has largely gone unnoticed and unexploited, despite Fisher discovering it almost a century ago. To make practical use of the transformation, I developed an heuristic Gaussian-based method that gave simple and accurate solutions including genetic drift, selection and mutation. This work (Khatri, Sci. Rep. 2016) is a key foundational basis for analysing longitudinal deep-sequencing data. More recently, I used the same framework for a new analysis which can distinguish between transmission and intrahost fitness effects in phylogenetic trees (Khatri et al, in preparation) and a new method to calculate the rate of fixation of rare variants (Khatri, bioRxiv123232, 2017).
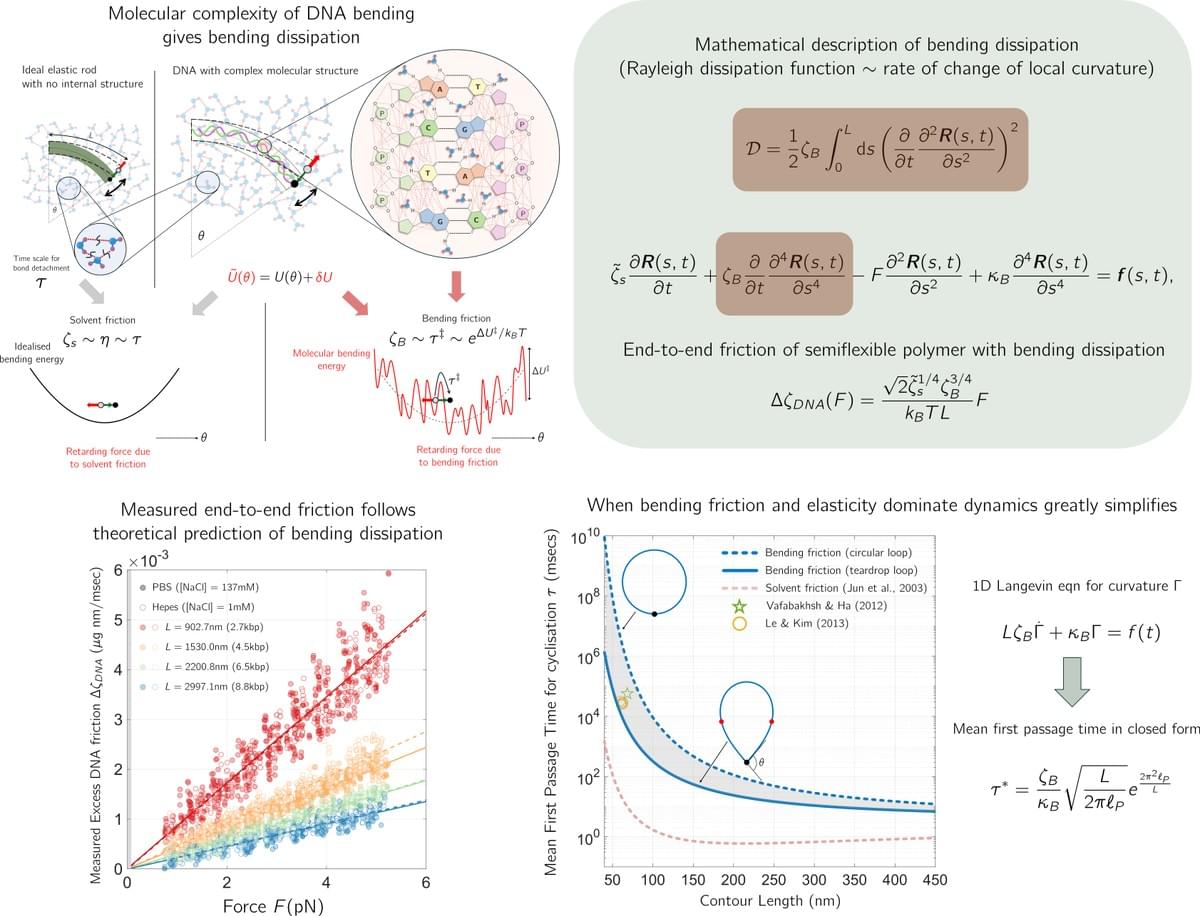
New polymer physics of DNA bending dissipation
A new material constant of DNA
DNA bending and looping is crucial for gene expression, packaging, and chromatin organisation. Yet current theories of DNA dynamics based on the semiflexible polymers underestimate empirically measured DNA loop-closure times by a remarkable 1000-fold. We have lacked a basic understanding of what determines how quickly DNA bends? With experimental biophysicist Maxim Moldtsov (Francis Crick Institute and Department of Physics, UCL), we have discovered that for lengths of DNA between 50 nm and 400 nm it is not solvent friction — as semiflexible polymer theories assume — but a new material constant of DNA, the bending friction, that determines how quickly DNA responds to mechanical forces. We determine this new material constant of DNA — from single molecule stretching experiments using optical tweezers — to be ζB = 241±17 μg/msec and strikingly independent of changing salt concentration (unlike the bending elasticity). This reveals a new picture of DNA mechanics at these length scales: rather than an ideal elastic rod, DNA is a viscoelastic polymer with a locally complex energy landscape for bending with dramatically slower dynamics. This is a fundamental discovery on par with establishing DNA’s elastic properties, which are well-described by the Worm-Like Chain model. While the bending elasticity has been profoundly influential, it is likely that bending friction and the Dissipative WLC (DWLC) model will be even more so: molecular biology is inherently out-of-equilibrium. This work has established a new foundation for DNA dynamics: any biological process involving DNA looping — from the action of molecular machines to chromosome organisation —
must account for this friction.
Publications
- George Pobegalov, Maxim Molodtsov, Bhavin S. Khatri, A new material constant of DNA is the major determinant of looping dynamics. bioRxiv doi.org/10.1101/2025.07.10.664086.
- Ioanna Morianou, Andrew Hammond, Bhavin S. Khatri, Austin Burt, Tony Nolan, & Andrea Crisanti, Engineering Resilient Gene Drives Towards Sustainable Malaria Control: Predicting, Testing and Overcoming Target Site Resistance. bioRxiv: doi.org/10.1101/2024.10.21.618489.
- Bhavin S. Khatri, A theory of the dynamics of DNA loop initiation in condensin/cohesin complexes, bioRxiv doi.org/10.1101/2021.11.10.468125.
- Yasu Kakui, Yoshiharu Kusano, Tereza Clarence, Maya Lopez, Todd Fallesen, Toru Hirota, Bhavin S Khatri, Frank Uhlmann, Progressive chromosome shape changes during cell divisions. EMBO Rep. 1–17 (2025).
- Bhavin S. Khatri, A simple stochastic theory of extinction shows rapid elimination of a Sars-like pandemic. Eur. Lett. (2025)
- Bhavin S. Khatri & Austin Burt, A theory of resistance to multiplexed gene drive demonstrates the significant role of weakly deleterious natural genetic variation, Proceeding of the National Academy of Sciences, 119, 32, e2200567119 (2022)
- From genotypes to organisms: State-of-the-art and perspectives of a cornerstone in evolutionary dynamics, Susanna Manrubia, José A. Cuesta, Jacobo Aguirre, Sebastian E. Ahnert, Lee Altenberg, Alejandro V. Cano, Pablo Catalán, Ramon Diaz-Uriarte, Santiago F. Elena, Juan Antonio García-Martín, Paulien Hogeweg, Bhavin S. Khatri, Joachim Krug, Ard A. Louis, Nora S. Martin, Joshua L. Payne, Matthew J. Tarnowski, Marcel Weiß, Physics of Life Reviews, 38, Pages 55-106 (2021)
- Computational and experimental analysis of mitotic chromosome formation pathways in fission yeast, Tereza Gerguri, Xiao Fu, Yasutaka Kakui, Bhavin S. Khatri, Christopher Barrington, Paul A. Bates, and Frank Uhlmann, Nucleic Acids Research, 9, p1294-1312, (2021)
- Fission yeast condensin maintains interphase chromatin organization to prevent transcription-coupled DNA damage
Yasutaka Kakui; Christopher Barrington; David J Barry; Tereza Gerguri; Xiao Fu; Paul A Bates; Bhavin S Khatri, Frank Uhlmann. Genome Biology, 21:272 (2020). - Biophysics and population size constrains speciation in an evolutionary model of developmental system drift, B.S. Khatri and R. A. Goldstein, PLoS Computational Biology, 15(7): e1007177, https://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1007177, 2019
- Robust estimation of recent effective population size from number of independent origins in soft sweeps, B.S. Khatri & A. Burt, Molecular Biology and Evolution, msz081, doi:10.1093/molbev/msz081 2019
- Survival of the frequent at finite population size and mutation rate, B. S. Khatri, bioRxiv doi: https://doi.org/10.1101/375147 2018
- Rate of fixation of rare variants in a population, B. S. Khatri, bioRxiv doi:10.1101/123232 2017
- Optimal chemotactic responses in stochastic environments, M. Godany, B. S. Khatri and R.A. Goldstein, PLoS ONE, 12(6):e0179111, 2017
- Quantifying evolutionary dynamics from variant-frequency time series, B. S. Khatri, Scientific Reports, 6:32497, 2016.
- Simple Biophysical Model Predicts Faster Accumulation of Hybrid Incompatibilities in Small Populations Under Stabilizing Selection, B. S. Khatri and Richard A. Goldstein, Genetics, 201(4), 1525-1537, 2015.
- Multiple-line inference of selection on quantitative traits, N. Riedel, B. S. Khatri, M. Lässig and J. Berg, Genetics, 201(1), 305-322, 2015.
- A coarse-grained biophysical model of sequence evolution and the population size dependence of the speciation rate, B. S. Khatri and R. Goldstein, Journal of Theoretical Biology, 378, 56-64, 2015.
- Oscillating microbial dynamics driven by small populations, limited nutrient supply and high death rates, B. S. Khatri, A. Free and R. Allen, Journal of Theoretical Biology, 314, 120{129, 2012.
- Fluctuation power spectra reveal dynamical heterogeneity of peptides, B. S. Khatri, Z.T. Yew, T.C.B. McLeish, E. Paci, Journal of Chemical Physics, 133, 015101, 2010.
- Dynamics of the coiled-coil unfolding transition of myosin rod probed by dissipation force spectrum, Y. Taniguchi, B. S. Khatri, D. J. Brockwell, E. Paci and M. Kawakami, Biophysical Journal, 99, 257-262, 2010.
- Statistical Mechanics of Convergent Evolution in Spatial Patterning, B. S. Khatri, T.C.B. McLeish and R.P. Sear, PNAS, 106, 9564-9569, 2009.
- Internal Friction of Polypeptide Chains at High Stretch, B. S. Khatri, K. Byrne, M. Kawakami, D.J. Brockwell, D. A. Smith, S.E. Radford and T.C.B. McLeish, Faraday Discuss., 139, 35 - 51, 2008.
- Rouse Model with Internal Friction: A Coarse Grained Framework for Single Biopolymer Dynamics, B. S. Khatri and Tom C.B. McLeish, Macromolecules, 40 (18), 6770 -6777, 2007.
- Entropy and Barrier-Controlled Fluctuations Determine Conformational Viscoelasticity in Single Biomolecules, B.S. Khatri, M. Kawakami, K. Byrne, D.A. Smith, T.C.B. McLeish, Biophysical Journal, 92, 1825-1835, 2007.
- Viscoelastic Properties of Single Poly(ethylene glycol) Molecules, M. Kawakami, K. Byrne, B.S. Khatri, T.C.B. McLeish, and D.A. Smith, ChemPhysChem, 7, 1710 -1716, 2006.
- Viscoelastic Measurements of Single Molecules on a Millisecond Time Scale by Magnetically Driven Oscillation of an Atomic Force Microscope Cantilever, M. Kawakami, K.Byrne, B.S. Khatri, T.C.B. McLeish, S.E. Radford and D.A. Smith, Langmuir, 21, 4765-4772, 2005.
- Viscoelastic Properties of Single Polysaccharide Molecules Determined by Analysis of Thermally Driven Oscillations of an Atomic Force Microscope Cantilever, M. Kawakami, K. Byrne, B.S. Khatri, T.C.B. McLeish, S.E. Radford and D.A. Smith, Langmuir, 401, 400-403, 2004.
Contact


Email
bhavin.khatri@
physics.org 
Research Fellow in Statistical Genomics
Imperial College London
Department of Life Sciences
Silwood Park
&
Visiting Scientist
The Francis Crick Institute
London





